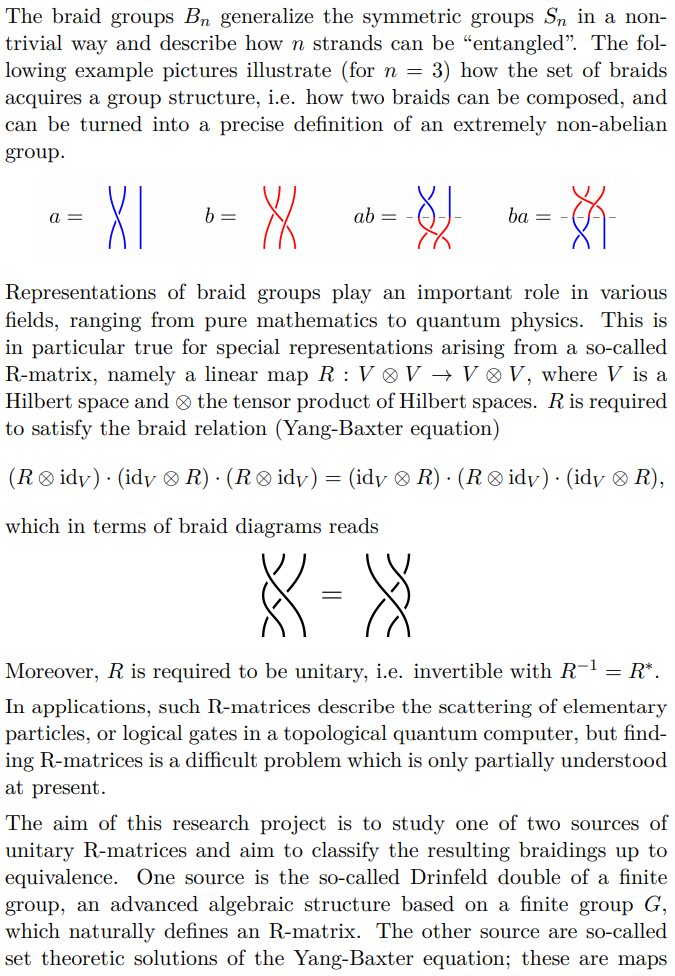
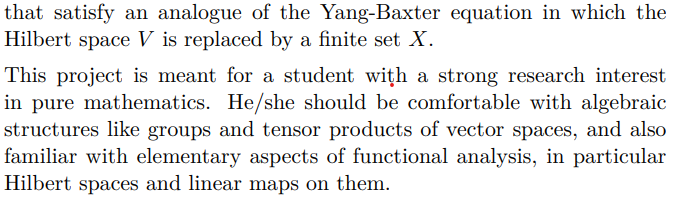
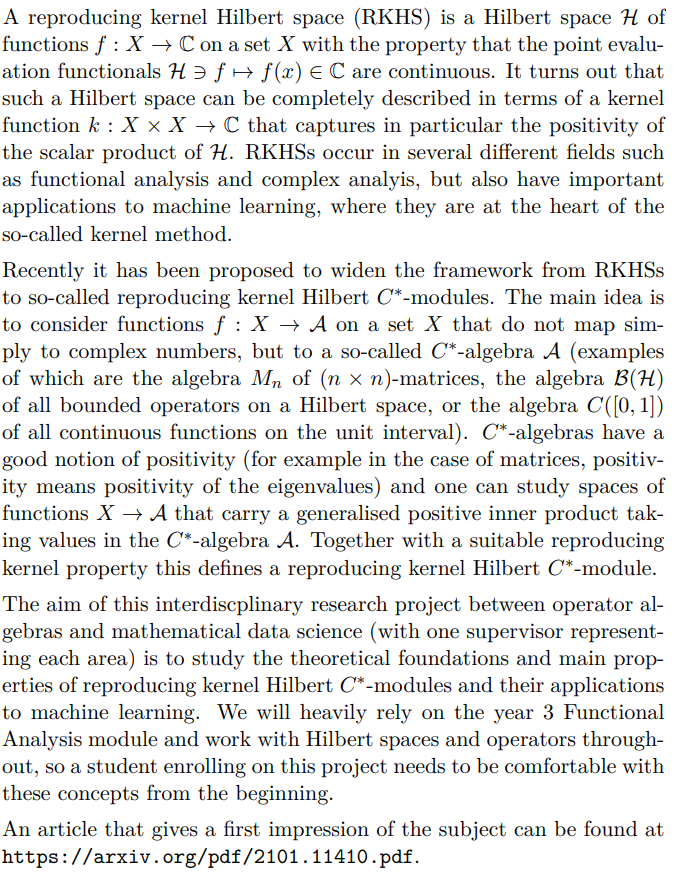
Title of Project: Dimension Reduction and Classification Algorithms
Code: AA2122A
Supervisor: Dr A. Artemiou
Project description:
Data mining and dimension reduction are two topics which receive great attention in the big data era we are in. There are a number of algorithms which were proposed to discuss data mining algorithms (like Support Vector Machines) and dimension reduction algorithms (like Sufficient Dimension Reduction). Recently, the two areas were combined to create new algorithms for dimension reduction in regression using ideas from the data mining literature. This created a class of new algorithms that perform very well and are able to do linear and nonlinear dimension reduction in a unified framework.
The project objective is to create first new algorithms for data mining by combining existing algorithms. The student will review the given literature and then derive results for new methodology for classification. The performance of the new algorithm will be evaluated using real datasets. Then, the algorithms will be used to perform dimension reduction to regression problems based on existing literature. Good programming skills are essential for this project. (In 2017-2018 a student work on this project and developed a modified version of SVM)
Prerequisite 3rd year module:
MA2500: Foundations of Probability and Statistics;
MA 2501: Programming and Statistics
and MA3505: Multivariate Statistics is helpful but not essential.
Number of students who can be supervised on this project:
1